#include <linerecord.h>
Public Member Functions | |
| LineRecord () | |
| Default constructor. More... | |
| LineRecord (Index species, Index isotopologue, Numeric f, Numeric psf, Numeric i0, Numeric ti0, Numeric elow, Numeric agam, Numeric sgam, Numeric nair, Numeric nself, Numeric, const ArrayOfNumeric &, Numeric, Numeric, Numeric, Numeric, Numeric, Numeric, Numeric) | |
| Constructor that sets all data elements explicitly. More... | |
| String | VersionString () const |
| Return the version String. More... | |
| Index | Version () const |
| Return the version number. More... | |
| void | SetVersion5 () |
| void | SetVersionToLatest () |
| Index | Species () const |
| The index of the molecular species that this line belongs to. More... | |
| Index | Isotopologue () const |
| The index of the isotopologue species that this line belongs to. More... | |
| String | Name () const |
| The full name of the species and isotopologue. More... | |
| const SpeciesRecord & | SpeciesData () const |
| The matching SpeciesRecord from species_data. More... | |
| const IsotopologueRecord & | IsotopologueData () const |
| The matching IsotopologueRecord from species_data. More... | |
| Numeric | F () const |
| The line center frequency in Hz. More... | |
| void | setF (Numeric new_mf) |
| Set the line center frequency in Hz. More... | |
| Numeric | Psf () const |
| The pressure shift parameter in Hz/Pa. More... | |
| Numeric | I0 () const |
The line intensity in m^2*Hz at the reference temperature Ti0. More... | |
| void | setI0 (Numeric new_mi0) |
| Set Intensity. More... | |
| Numeric | Ti0 () const |
| Reference temperature for I0 in K: More... | |
| Numeric | Elow () const |
| Lower state energy in cm^-1: More... | |
| void | SetElow (Numeric new_melow) |
| Numeric | Evlow () const |
| Lower state vibrational energy in cm^-1: More... | |
| void | SetEvlow (Numeric evlow) |
| Index | NLTELowerIndex () const |
| void | SetNLTELowerIndex (Index nlte_lower_index) |
| Numeric | Evupp () const |
| Upper state vibrational energy in cm^-1: More... | |
| void | SetEvupp (Numeric evupp) |
| Index | NLTEUpperIndex () const |
| void | SetNLTEUpperIndex (Index nlte_upper_index) |
| Numeric | Agam () const |
| Air broadened width in Hz/Pa: More... | |
| Numeric | Sgam () const |
| Self broadened width in Hz/Pa: More... | |
| Numeric | Nair () const |
| AGAM temperature exponent (dimensionless): More... | |
| Numeric | Nself () const |
| SGAM temperature exponent (dimensionless): More... | |
| Numeric | A () const |
| ARTSCAT-4/5 Einstein A-coefficient in 1/s : More... | |
| Numeric | electric_dipole_moment_squared () const |
| Numeric | magnetic_quadrapole_moment_squared () const |
| Numeric | reduced_rovibrational_dipole (Rational k=1) const |
| Numeric | reduced_magnetic_quadrapole () const |
| Numeric | G_upper () const |
| ARTSCAT-4/5 Upper state stat. More... | |
| Numeric | G_lower () const |
| ARTSCAT-4/5 Lower state stat. More... | |
| void | SetQuantumNumberLower (const Index i, const Rational r) |
| Quantum numbers. More... | |
| void | SetQuantumNumberLower (const String i, const Rational r) |
| void | SetQuantumNumberLower (const QuantumNumberType i, const Rational r) |
| void | SetQuantumNumberUpper (const QuantumNumberType i, const Rational r) |
| void | SetQuantumNumberUpper (const String i, const Rational r) |
| void | SetQuantumNumberUpper (const Index i, const Rational r) |
| const QuantumIdentifier & | QuantumIdentity () const |
| Quantum identifier. More... | |
| const QuantumNumbers & | LowerQuantumNumbers () const |
| const QuantumNumbers & | UpperQuantumNumbers () const |
| Rational | LowerQuantumNumber (QuantumNumberType X) const |
| Rational | UpperQuantumNumber (QuantumNumberType X) const |
| bool | InQuantumID (const QuantumIdentifier &qid) const |
| bool | UpperStateInQuantumID (const QuantumIdentifier &qid) const |
| bool | LowerStateInQuantumID (const QuantumIdentifier &qid) const |
| bool | do_linemixing (const Numeric &P) const noexcept |
| Do linemixing test. More... | |
| void | SetLineMixingParameters (Numeric &Y, Numeric &G, Numeric &DV, const Numeric &T, const Numeric &P, const ConstVectorView vmrs, const ArrayOfArrayOfSpeciesTag &abs_species) const |
| void | SetPressureBroadeningParameters (Numeric &G0, Numeric &G2, Numeric &ETA, Numeric &D0, Numeric &D2, Numeric &FVC, const Numeric &T, const Numeric &P, const ConstVectorView vmrs, const ArrayOfArrayOfSpeciesTag &abs_species) const |
| void | SetPressureBroadeningParametersTemperatureDerivative (Numeric &dG0, Numeric &dG2, Numeric &dETA, Numeric &dD0, Numeric &dD2, Numeric &dFVC, const Numeric &T, const Numeric &P, const ConstVectorView vmrs, const ArrayOfArrayOfSpeciesTag &abs_species) const |
| void | SetLineMixing2SecondOrderData (ConstVectorView d) |
| void | SetLineMixing2AER (ConstVectorView d) |
| LineShape::Model & | GetLineShapeModel () |
| const LineShape::Model & | GetLineShapeModel () const |
| void | SetLineShapeModel (const LineShape::Model &lsm) |
| void | LineShapeModelOnlyAir () |
| void | LineShapeModelRemoveSelf () |
| void | SetLineShapeModelParameters (const LineShape::ModelParameters &x, const String &spec, const String &var) |
| void | SetLineShapeModelParameter (const Numeric &X, const String &spec, const String &var, const String &coeff) |
| LineShape::ModelParameters | GetLineShapeModelParameters (const String &spec, const String &var) const |
| void | SetSpecial () |
| void | SetStandard () |
| bool | LineMixingByBand () const |
| bool | LineShapeModelHasAir () const noexcept |
| void | SetAirPressureBroadening (Numeric &G0, Numeric &D0, const Numeric &T, const Numeric &P, const Numeric &self_vmr) const |
| Numeric | PressureBroadeningAirBroadeningNair () const |
| Numeric | PressureBroadeningAirBroadeningPsf () const |
| Numeric | PressureBroadeningAirBroadeningAgam () const |
| Numeric | GetPrepInternalDerivative (const Numeric &T, const Numeric &P, const Vector &vmrs, const RetrievalQuantity &derivative) const noexcept |
| Numeric | GetInternalDerivative (const Numeric &T, const Numeric &P, const ConstVectorView vmrs, const ArrayOfArrayOfSpeciesTag &abs_species, const RetrievalQuantity &derivative) const |
| LineShape::Output | GetPrepShapeParams (const Numeric &T, const Numeric &P, const Vector &vmrs) const noexcept |
| LineShape::Output | GetShapeParams (const Numeric &T, const Numeric &P, const ConstVectorView vmrs, const ArrayOfArrayOfSpeciesTag &abs_species) const |
| LineShape::Output | GetPrepShapeParams_dT (const Numeric &T, const Numeric &P, const Vector &vmrs) const noexcept |
| LineShape::Output | GetShapeParams_dT (const Numeric &T, const Numeric &P, const ConstVectorView vmrs, const ArrayOfArrayOfSpeciesTag &abs_species) const |
| LineShape::Output | GetShapeParams_dVMR (const Numeric &T, const Numeric &P, const QuantumIdentifier &vmr_qi) const noexcept |
| void | ZeemanModelInit () |
| Zeeman Effect Data. More... | |
| void | ZeemanModelInitZero () |
| Numeric | ZeemanModelStrength (Zeeman::Polarization type, Index i) const |
| Numeric | ZeemanModelSplitting (Zeeman::Polarization type, Index i) const |
| Index | ZeemanModelLineCount (Zeeman::Polarization type) const |
| Zeeman::Model | ZeemanModel () const |
| Zeeman::Model & | ZeemanModel () |
| const Numeric & | CutOff () const |
| Cutoff frequency. More... | |
| void | SetCutOff (const Numeric &cutoff) |
| const Numeric & | LineMixingLimit () const |
| Line mixing pressure limit. More... | |
| void | SetLineMixingLimit (const Numeric &limit) |
| const MirroringType & | GetMirroringType () const |
| Line shape mirroring factor. More... | |
| void | SetMirroringType (const MirroringType in) |
| String | GetMirroringTypeString () const |
| const LineNormalizationType & | GetLineNormalizationType () const |
| Line shape normalization factor. More... | |
| void | SetLineNormalizationType (const LineNormalizationType in) |
| String | GetLineNormalizationTypeString () const |
| LineShape::Type | GetLineShapeType () const noexcept |
| const LinePopulationType & | GetLinePopulationType () const |
| Line population type. More... | |
| void | SetLinePopulationType (const LinePopulationType in) |
| String | GetLinePopulationTypeString () const |
| bool | IsNotSameSpecIso (const LineRecord &other) const |
| bool | ReadFromHitran2001Stream (istream &is, const Verbosity &verbosity) |
| Read one line from a stream associated with a HITRAN 1986-2001 file. More... | |
| bool | ReadFromLBLRTMStream (istream &is, const Verbosity &verbosity) |
| LBLRTM uses the same format as HITRAN pre-2004 but also carry line mixing data, so we must read it separately. More... | |
| bool | ReadFromHitran2004Stream (istream &is, const Verbosity &verbosity, const Numeric fmin=0) |
| Read one line from a stream associated with a HITRAN 2004 file. More... | |
| bool | ReadFromMytran2Stream (istream &is, const Verbosity &verbosity) |
| Read one line from a stream associated with a MYTRAN2 file. More... | |
| bool | ReadFromJplStream (istream &is, const Verbosity &verbosity) |
| Read one line from a stream associated with a JPL file. More... | |
| bool | ReadFromArtscat3Stream (istream &is, const Verbosity &verbosity) |
| Read one line from a stream associated with an ARTSCAT-3 file. More... | |
| bool | ReadFromArtscat4Stream (istream &is, const Verbosity &verbosity) |
| Read one line from a stream associated with an ARTSCAT-4 file. More... | |
| bool | ReadFromArtscat5Stream (istream &is, const Verbosity &verbosity) |
| Read one line from a stream associated with an ARTSCAT-5 file. More... | |
Private Attributes | |
| Index | mversion |
| QuantumIdentifier | mqid |
| Numeric | mf |
| Numeric | mi0 |
| Numeric | mti0 |
| Numeric | melow |
| Numeric | mevlow |
| Index | mnlte_lower_index |
| Numeric | mevupp |
| Index | mnlte_upper_index |
| Numeric | ma |
| Numeric | mgupper |
| Numeric | mglower |
| LineShape::Model | mlineshapemodel |
| Line function data (pressure broadening and line mixing) More... | |
| bool | mstandard |
| FIXME Richard, please explain. More... | |
| Zeeman::Model | mzeemanmodel |
| Zeeman effect model class. More... | |
| Numeric | mcutoff |
| Cutoff frequency. More... | |
| Numeric | mlinemixing_limit |
| Line mixing pressure limit. More... | |
| MirroringType | mmirroring |
| Line shape mirroring effect type. More... | |
| LineNormalizationType | mlinenorm |
| Line shape normalization type. More... | |
| LinePopulationType | mpopulation |
| Line LTE/NLTE type. More... | |
Detailed Description
Definition at line 228 of file linerecord.h.
Constructor & Destructor Documentation
◆ LineRecord() [1/2]
|
inline |
Default constructor.
Initialize to default values. The indices are initialized to large numbers, so that we at least get range errors when we try to used un-initialized data.
Definition at line 233 of file linerecord.h.
◆ LineRecord() [2/2]
|
inline |
Constructor that sets all data elements explicitly.
If assertions are not disabled (i.e., if NDEBUG is not #defined), assert statements check that the species and isotopologue data exists.
Definition at line 259 of file linerecord.h.
Member Function Documentation
◆ A()
|
inline |
ARTSCAT-4/5 Einstein A-coefficient in 1/s :
Definition at line 410 of file linerecord.h.
Referenced by operator<<().
◆ Agam()
|
inline |
Air broadened width in Hz/Pa:
Definition at line 398 of file linerecord.h.
◆ CutOff()
|
inline |
◆ do_linemixing()
|
inlinenoexcept |
Do linemixing test.
Definition at line 497 of file linerecord.h.
◆ electric_dipole_moment_squared()
|
inline |
Definition at line 412 of file linerecord.h.
◆ Elow()
|
inline |
Lower state energy in cm^-1:
Definition at line 378 of file linerecord.h.
Referenced by operator<<().
◆ Evlow()
|
inline |
Lower state vibrational energy in cm^-1:
Definition at line 382 of file linerecord.h.
◆ Evupp()
|
inline |
Upper state vibrational energy in cm^-1:
Definition at line 390 of file linerecord.h.
◆ F()
|
inline |
The line center frequency in Hz.
Definition at line 349 of file linerecord.h.
Referenced by operator<<().
◆ G_lower()
|
inline |
ARTSCAT-4/5 Lower state stat.
weight:
Definition at line 452 of file linerecord.h.
Referenced by operator<<().
◆ G_upper()
|
inline |
ARTSCAT-4/5 Upper state stat.
weight:
Definition at line 449 of file linerecord.h.
Referenced by operator<<().
◆ GetInternalDerivative()
|
inline |
Definition at line 672 of file linerecord.h.
◆ GetLineNormalizationType()
|
inline |
Line shape normalization factor.
Definition at line 789 of file linerecord.h.
Referenced by operator<<().
◆ GetLineNormalizationTypeString()
| String LineRecord::GetLineNormalizationTypeString | ( | ) | const |
Definition at line 2833 of file linerecord.cc.
References mlinenorm, None, RosenkranzQuadratic, VVH, and VVW.
Referenced by operator<<().
◆ GetLinePopulationType()
|
inline |
Line population type.
Definition at line 802 of file linerecord.h.
◆ GetLinePopulationTypeString()
| String LineRecord::GetLinePopulationTypeString | ( | ) | const |
Definition at line 2861 of file linerecord.cc.
References ByLTE, ByPopulationDistribution, ByVibrationalTemperatures, and mpopulation.
◆ GetLineShapeModel() [1/2]
|
inline |
Definition at line 573 of file linerecord.h.
Referenced by operator<<().
◆ GetLineShapeModel() [2/2]
|
inline |
Definition at line 574 of file linerecord.h.
◆ GetLineShapeModelParameters()
|
inline |
Definition at line 606 of file linerecord.h.
References LineShape::string2variable().
◆ GetLineShapeType()
|
inlinenoexcept |
Definition at line 797 of file linerecord.h.
◆ GetMirroringType()
|
inline |
Line shape mirroring factor.
Definition at line 784 of file linerecord.h.
Referenced by operator<<().
◆ GetMirroringTypeString()
| String LineRecord::GetMirroringTypeString | ( | ) | const |
Definition at line 2803 of file linerecord.cc.
References Lorentz, Manual, mmirroring, None, and SameAsLineShape.
Referenced by operator<<().
◆ GetPrepInternalDerivative()
|
inlinenoexcept |
Definition at line 641 of file linerecord.h.
References LineShape::vmrs().
◆ GetPrepShapeParams()
|
inlinenoexcept |
Definition at line 681 of file linerecord.h.
References LineShape::vmrs(), and LineShape::Output::Y.
◆ GetPrepShapeParams_dT()
|
inlinenoexcept |
Definition at line 699 of file linerecord.h.
References LineShape::vmrs(), and LineShape::Output::Y.
◆ GetShapeParams()
|
inline |
Method to compute the line mixing and pressure broadening parameters
Definition at line 690 of file linerecord.h.
◆ GetShapeParams_dT()
|
inline |
Method to compute the temperature derivatives of line mixing and pressure broadening parameters
Definition at line 708 of file linerecord.h.
◆ GetShapeParams_dVMR()
|
inlinenoexcept |
Method to compute the temperature derivatives of line mixing and pressure broadening parameters
Definition at line 718 of file linerecord.h.
References LineShape::differenceOutput(), and LineShape::Output::Y.
◆ I0()
|
inline |
The line intensity in m^2*Hz at the reference temperature Ti0.
The line intensity 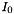 is defined by:
is defined by:
![\[ \alpha(\nu) = n \, x \, I_0(T) \, F(\nu) \]](form_6.png)
where 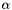 is the absorption coefficient (in m^-1),
is the absorption coefficient (in m^-1), 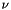 is frequency,
is frequency, 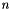 is the total number density,
is the total number density, 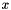 is the volume mixing ratio, and
is the volume mixing ratio, and 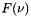 is the lineshape function.
is the lineshape function.
Definition at line 369 of file linerecord.h.
Referenced by operator<<().
◆ InQuantumID()
|
inline |
Definition at line 488 of file linerecord.h.
◆ IsNotSameSpecIso()
|
inline |
Definition at line 808 of file linerecord.h.
References QuantumIdentifier::Isotopologue(), mqid, Absorption::ReadFromArtscat3Stream(), Absorption::ReadFromArtscat4Stream(), Absorption::ReadFromArtscat5Stream(), Absorption::ReadFromHitran2001Stream(), Absorption::ReadFromHitran2004Stream(), Absorption::ReadFromJplStream(), Absorption::ReadFromLBLRTMStream(), Absorption::ReadFromMytran2Stream(), and QuantumIdentifier::Species().
◆ Isotopologue()
|
inline |
The index of the isotopologue species that this line belongs to.
The isotopologue species data can be accessed by species_data[Species()].Isotopologue()[Isotopologue()].
Definition at line 317 of file linerecord.h.
◆ IsotopologueData()
| const IsotopologueRecord & LineRecord::IsotopologueData | ( | ) | const |
The matching IsotopologueRecord from species_data.
The IsotopologueRecord is a subset of the SpeciesRecord. To get at the isotopologue data of a LineRecord lr, you can use:
- species_data[lr.Species()].Isotopologue()[lr.Isotopologue()]
- lr.SpeciesData().Isotopologue()[lr.Isotopologue()]
- lr.IsotopologueData()
The last option is clearly the shortest, and has the advantage that you don't have to declare the external variable species_data.
Definition at line 55 of file linerecord.cc.
References QuantumIdentifier::Isotopologue(), mqid, QuantumIdentifier::Species(), and global_data::species_data.
◆ LineMixingByBand()
|
inline |
Definition at line 614 of file linerecord.h.
◆ LineMixingLimit()
|
inline |
Line mixing pressure limit.
Definition at line 780 of file linerecord.h.
Referenced by operator<<().
◆ LineShapeModelHasAir()
|
inlinenoexcept |
Definition at line 616 of file linerecord.h.
◆ LineShapeModelOnlyAir()
|
inline |
Definition at line 578 of file linerecord.h.
◆ LineShapeModelRemoveSelf()
|
inline |
Definition at line 587 of file linerecord.h.
◆ LowerQuantumNumber()
|
inline |
Definition at line 482 of file linerecord.h.
◆ LowerQuantumNumbers()
|
inline |
Definition at line 476 of file linerecord.h.
Referenced by operator<<().
◆ LowerStateInQuantumID()
|
inline |
Definition at line 492 of file linerecord.h.
◆ magnetic_quadrapole_moment_squared()
|
inline |
Definition at line 417 of file linerecord.h.
◆ Nair()
|
inline |
AGAM temperature exponent (dimensionless):
Definition at line 404 of file linerecord.h.
◆ Name()
| String LineRecord::Name | ( | ) | const |
The full name of the species and isotopologue.
E.g., `O3-666'. The name is found by looking up the information in species_data, using the species and isotopologue index.
Definition at line 42 of file linerecord.cc.
References SpeciesRecord::Isotopologue(), QuantumIdentifier::Isotopologue(), mqid, SpeciesRecord::Name(), QuantumIdentifier::Species(), and global_data::species_data.
Referenced by operator<<().
◆ NLTELowerIndex()
|
inline |
Definition at line 384 of file linerecord.h.
Referenced by nlteSetByQuantumIdentifiers().
◆ NLTEUpperIndex()
|
inline |
Definition at line 392 of file linerecord.h.
Referenced by nlteSetByQuantumIdentifiers().
◆ Nself()
|
inline |
SGAM temperature exponent (dimensionless):
Definition at line 407 of file linerecord.h.
◆ PressureBroadeningAirBroadeningAgam()
|
inline |
Definition at line 639 of file linerecord.h.
◆ PressureBroadeningAirBroadeningNair()
|
inline |
Definition at line 637 of file linerecord.h.
◆ PressureBroadeningAirBroadeningPsf()
|
inline |
Definition at line 638 of file linerecord.h.
◆ Psf()
|
inline |
The pressure shift parameter in Hz/Pa.
Definition at line 355 of file linerecord.h.
◆ QuantumIdentity()
|
inline |
Quantum identifier.
Definition at line 475 of file linerecord.h.
Referenced by nlteSetByQuantumIdentifiers().
◆ ReadFromArtscat3Stream()
| bool LineRecord::ReadFromArtscat3Stream | ( | istream & | is, |
| const Verbosity & | verbosity | ||
| ) |
Read one line from a stream associated with an ARTSCAT-3 file.
Format: see Documentation of class LineRecord
The function attempts to read a line of data from the catalogue. It returns false if it succeeds. Otherwise, if eof is reached, it returns true. If an error occurs, a runtime_error is thrown. When the function looks for a data line, comment lines are automatically skipped.
- Parameters
-
is Stream from which to read
- Exceptions
-
runtime_error Some error occured during the read
- Returns
- false=ok (data returned), true=eof (no data returned)
- Date
- 2000-12-15
- Date
- 2001-06-20
Definition at line 2069 of file linerecord.cc.
References CREATE_OUT3, i, SpeciesRecord::Isotopologue(), mversion, SpeciesRecord::Name(), my_basic_string< charT >::nelem(), Array< base >::nelem(), global_data::species_data, and SpecIsoMap::Speciesindex().
◆ ReadFromArtscat4Stream()
| bool LineRecord::ReadFromArtscat4Stream | ( | istream & | is, |
| const Verbosity & | verbosity | ||
| ) |
Read one line from a stream associated with an ARTSCAT-4 file.
Format: see Documentation of class LineRecord
The function attempts to read a line of data from the catalogue. It returns false if it succeeds. Otherwise, if eof is reached, it returns true. If an error occurs, a runtime_error is thrown. When the function looks for a data line, comment lines are automatically skipped.
- Parameters
-
is Stream from which to read
- Exceptions
-
runtime_error Some error occured during the read
- Returns
- false=ok (data returned), true=eof (no data returned)
- Date
- 2012-02-10
Definition at line 2253 of file linerecord.cc.
References CREATE_OUT3, i, SpeciesRecord::Isotopologue(), mversion, SpeciesRecord::Name(), my_basic_string< charT >::nelem(), Array< base >::nelem(), global_data::species_data, and SpecIsoMap::Speciesindex().
◆ ReadFromArtscat5Stream()
| bool LineRecord::ReadFromArtscat5Stream | ( | istream & | is, |
| const Verbosity & | verbosity | ||
| ) |
Read one line from a stream associated with an ARTSCAT-5 file.
Format: see Documentation of class LineRecord
The function attempts to read a line of data from the catalogue. It returns false if it succeeds. Otherwise, if eof is reached, it returns true. If an error occurs, a runtime_error is thrown. When the function looks for a data line, comment lines are automatically skipped.
- Parameters
-
is Stream from which to read
- Exceptions
-
runtime_error Some error occured during the read
- Returns
- false=ok (data returned), true=eof (no data returned)
- Date
- 2014-12-01
Definition at line 2458 of file linerecord.cc.
References CREATE_OUT3, i, SpeciesRecord::Isotopologue(), mversion, SpeciesRecord::Name(), my_basic_string< charT >::nelem(), Array< base >::nelem(), global_data::species_data, and SpecIsoMap::Speciesindex().
◆ ReadFromHitran2001Stream()
| bool LineRecord::ReadFromHitran2001Stream | ( | istream & | is, |
| const Verbosity & | verbosity | ||
| ) |
Read one line from a stream associated with a HITRAN 1986-2001 file.
The HITRAN format is as follows (directly from the HITRAN documentation):
Each line consists of 100
bytes of ASCII text data, followed by a line feed (ASCII 10) and
carriage return (ASCII 13) character, for a total of 102 bytes per line.
Each line can be read using the following READ and FORMAT statement pair
(for a FORTRAN sequential access read):
READ(3,800) MO,ISO,V,S,R,AGAM,SGAM,E,N,d,V1,V2,Q1,Q2,IERF,IERS,
* IERH,IREFF,IREFS,IREFH
800 FORMAT(I2,I1,F12.6,1P2E10.3,0P2F5.4,F10.4,F4.2,F8.6,2I3,2A9,3I1,3I2)
Each item is defined below, with its format shown in parenthesis.
MO (I2) = molecule number
ISO (I1) = isotopologue number (1 = most abundant, 2 = second, etc)
V (F12.6) = frequency of transition in wavenumbers (cm-1)
S (E10.3) = intensity in cm-1/(molec * cm-2) at 296 Kelvin
R (E10.3) = transition probability squared in Debyes**2
AGAM (F5.4) = air-broadened halfwidth (HWHM) in cm-1/atm at 296 Kelvin
SGAM (F5.4) = self-broadened halfwidth (HWHM) in cm-1/atm at 296 Kelvin
E (F10.4) = lower state energy in wavenumbers (cm-1)
N (F4.2) = coefficient of temperature dependence of air-broadened halfwidth
d (F8.6) = shift of transition due to pressure (cm-1)
V1 (I3) = upper state global quanta index
V2 (I3) = lower state global quanta index
Q1 (A9) = upper state local quanta
Q2 (A9) = lower state local quanta
IERF (I1) = accuracy index for frequency reference
IERS (I1) = accuracy index for intensity reference
IERH (I1) = accuracy index for halfwidth reference
IREFF (I2) = lookup index for frequency
IREFS (I2) = lookup index for intensity
IREFH (I2) = lookup index for halfwidth
The molecule numbers are encoded as shown in the table below:
0= Null 1= H2O 2= CO2 3= O3 4= N2O 5= CO
6= CH4 7= O2 8= NO 9= SO2 10= NO2 11= NH3
12= HNO3 13= OH 14= HF 15= HCl 16= HBr 17= HI
18= ClO 19= OCS 20= H2CO 21= HOCl 22= N2 23= HCN
24=CH3Cl 25= H2O2 26= C2H2 27= C2H6 28= PH3 29= COF2
30= SF6 31= H2S 32=HCOOH
The function attempts to read a line of data from the catalogue. It returns false if it succeeds. Otherwise, if eof is reached, it returns true. If an error occurs, a runtime_error is thrown. When the function looks for a data line, comment lines are automatically skipped. It is checked if the data record has the right number of characters. If not, a runtime_error is thrown.
- Parameters
-
is Stream from which to read
- Exceptions
-
runtime_error Some error occured during the read
- Returns
- false=ok (data returned), true=eof (no data returned)
Definition at line 61 of file linerecord.cc.
References CREATE_OUT0, CREATE_OUT3, extract(), i, iso(), SpeciesRecord::Isotopologue(), QuantumIdentifier::Isotopologue(), max, mf, mqid, my_basic_string< charT >::nelem(), Array< base >::nelem(), Absorption::nelem(), QuantumIdentifier::Species(), global_data::species_data, SPEED_OF_LIGHT, and _CS_basic_sstream_base< _CS_cT, _CS_Tr, _CS_Al >::str().
◆ ReadFromHitran2004Stream()
| bool LineRecord::ReadFromHitran2004Stream | ( | istream & | is, |
| const Verbosity & | verbosity, | ||
| const Numeric | fmin = 0 |
||
| ) |
Read one line from a stream associated with a HITRAN 2004 file.
The HITRAN format is as follows:
Each line consists of 160 ASCII characters, followed by a line feed (ASCII 10) and carriage return (ASCII 13) character, for a total of 162 bytes per line. Each item is defined below, with its Fortran format shown in parenthesis. (I2) molecule number (I1) isotopologue number (1 = most abundant, 2 = second, etc) (F12.6) vacuum wavenumbers (cm-1) (E10.3) intensity in cm-1/(molec * cm-2) at 296 Kelvin (E10.3) Einstein-A coefficient (s-1) (F5.4) air-broadened halfwidth (HWHM) in cm-1/atm at 296 Kelvin (F5.4) self-broadened halfwidth (HWHM) in cm-1/atm at 296 Kelvin (F10.4) lower state energy (cm-1) (F4.2) coefficient of temperature dependence of air-broadened halfwidth (F8.6) air-broadened pressure shift of line transition at 296 K (cm-1) (A15) upper state global quanta (A15) lower state global quanta (A15) upper state local quanta (A15) lower state local quanta (I1) uncertainty index for wavenumber (I1) uncertainty index for intensity (I1) uncertainty index for air-broadened half-width (I1) uncertainty index for self-broadened half-width (I1) uncertainty index for temperature dependence (I1) uncertainty index for pressure shift (I2) index for table of references correspond. to wavenumber (I2) index for table of references correspond. to intensity (I2) index for table of references correspond. to air-broadened half-width (I2) index for table of references correspond. to self-broadened half-width (I2) index for table of references correspond. to temperature dependence (I2) index for table of references correspond. to pressure shift (A1) flag (*) for lines supplied with line-coupling algorithm (F7.1) upper state statistical weight (F7.1) lower state statistical weight The molecule numbers are encoded as shown in the table below: 0= Null 1= H2O 2= CO2 3= O3 4= N2O 5= CO 6= CH4 7= O2 8= NO 9= SO2 10= NO2 11= NH3 12= HNO3 13= OH 14= HF 15= HCl 16= HBr 17= HI 18= ClO 19= OCS 20= H2CO 21= HOCl 22= N2 23= HCN 24=CH3Cl 25= H2O2 26= C2H2 27= C2H6 28= PH3 29= COF2 30= SF6 31= H2S 32=HCOOH 33= HO2 34= O 35=ClONO2 36= NO+ 37= HOBr 38= C2H4
CH3OH is not included in ARTS because its total internal partition sum is not known yet.
The function attempts to read a line of data from the catalogue. It returns false if it succeeds. Otherwise, if eof is reached, it returns true. If an error occurs, a runtime_error is thrown. When the function looks for a data line, comment lines are automatically skipped. It is checked if the data record has the right number of characters (comment lines are ignored). If not, a runtime_error is thrown. If the molecule is unknown to ARTS, a warning is prompted but the program continues (ignoring this record). For CH3OH this warning will be issued even when using the regular Hitran 2004 data base (see above). If the line center is below fmin, mf is set to -1 and the caller should ignore this line.
- Parameters
-
is Stream from which to read verbosity Verbosity fmin Skip line if mf < fmin
- Exceptions
-
runtime_error Some error occured during the read
- Returns
- false=ok (data returned), true=eof (no data returned)
Definition at line 1020 of file linerecord.cc.
References CREATE_OUT1, CREATE_OUT3, extract(), i, iso(), SpeciesRecord::Isotopologue(), QuantumIdentifier::Isotopologue(), max, mf, mqid, my_basic_string< charT >::nelem(), Array< base >::nelem(), Absorption::nelem(), QuantumIdentifier::Species(), global_data::species_data, SPEED_OF_LIGHT, and _CS_basic_sstream_base< _CS_cT, _CS_Tr, _CS_Al >::str().
◆ ReadFromJplStream()
| bool LineRecord::ReadFromJplStream | ( | istream & | is, |
| const Verbosity & | verbosity | ||
| ) |
Read one line from a stream associated with a JPL file.
The JPL format is as follows (directly taken from the jpl documentation):
The catalog line files are composed of 80-character lines, with one
line entry per spectral line. The format of each line is:
\label{lfmt}
\begin{tabular}{@{}lccccccccr@{}}
FREQ, & ERR, & LGINT, & DR, & ELO, & GUP, & TAG, & QNFMT, & QN${'}$, & QN${''}$\\
(F13.4, & F8.4, & F8.4, & I2, & F10.4, & I3, & I7, & I4, & 6I2, & 6I2)\\
\end{tabular}
\begin{tabular}{lp{4.5in}}
FREQ: & Frequency of the line in MHz.\\
ERR: & Estimated or experimental error of FREQ in MHz.\\
LGINT: &Base 10 logarithm of the integrated intensity
in units of \linebreak nm$^2$$\cdot$MHz at 300 K. (See Section 3 for
conversions to other units.)\\
DR: & Degrees of freedom in the rotational partition
function (0 for atoms, 2 for linear molecules, and 3 for nonlinear
molecules).\\
ELO: &Lower state energy in cm$^{-1}$ relative to the lowest energy
spin--rotation level in ground vibronic state.\\
GUP: & Upper state degeneracy.\\
TAG: & Species tag or molecular identifier.
A negative value flags that the line frequency has
been measured in the laboratory. The absolute value of TAG is then the
species tag and ERR is the reported experimental error. The three most
significant digits of the species tag are coded as the mass number of the
species, as explained above.\\
QNFMT: &Identifies the format of the quantum numbers
given in the field QN. These quantum number formats are given in Section 5
and are different from those in the first two editions of the catalog.\\
QN${'}$: & Quantum numbers for the upper state coded
according to QNFMT.\\
QN${''}$: & Quantum numbers for the lower state.\\
\end{tabular}
The function attempts to read a line of data from the catalogue. It returns false if it succeeds. Otherwise, if eof is reached, it returns true. If an error occurs, a runtime_error is thrown. When the function looks for a data line, comment lines are automatically skipped (unused in jpl).
- Parameters
-
is Stream from which to read
- Exceptions
-
runtime_error Some error occured during the read
- Returns
- false=ok (data returned), true=eof (no data returned)
- Date
- 01.11.00
Definition at line 1843 of file linerecord.cc.
References CREATE_OUT3, i, SpeciesRecord::Isotopologue(), Array< base >::nelem(), and global_data::species_data.
◆ ReadFromLBLRTMStream()
| bool LineRecord::ReadFromLBLRTMStream | ( | istream & | is, |
| const Verbosity & | verbosity | ||
| ) |
LBLRTM uses the same format as HITRAN pre-2004 but also carry line mixing data, so we must read it separately.
- Date
- 2014-09-24
Definition at line 466 of file linerecord.cc.
References CREATE_OUT0, CREATE_OUT3, extract(), i, iso(), SpeciesRecord::Isotopologue(), QuantumIdentifier::Isotopologue(), max, mf, mqid, my_basic_string< charT >::nelem(), Array< base >::nelem(), Absorption::nelem(), QuantumIdentifier::Species(), global_data::species_data, SPEED_OF_LIGHT, and _CS_basic_sstream_base< _CS_cT, _CS_Tr, _CS_Al >::str().
◆ ReadFromMytran2Stream()
| bool LineRecord::ReadFromMytran2Stream | ( | istream & | is, |
| const Verbosity & | verbosity | ||
| ) |
Read one line from a stream associated with a MYTRAN2 file.
The MYTRAN2 format is as follows (directly taken from the abs_my.c documentation):
The MYTRAN format is as follows (FORTRAN notation):
FORMAT(I2,I1,F13.4,1PE10.3,0P2F5.2,F10.4,2F4.2,F8.6,F6.4,2I3,2A9,4I1,3I2)
Each item is defined below, with its FORMAT String shown in
parenthesis.
MO (I2) = molecule number
ISO (I1) = isotopologue number (1 = most abundant, 2 = second, etc)
* F (F13.4) = frequency of transition in MHz
* errf (F8.4) = error in f in MHz
S (E10.3) = intensity in cm-1/(molec * cm-2) at 296 K
* AGAM (F5.4) = air-broadened halfwidth (HWHM) in MHz/Torr at Tref
* SGAM (F5.4) = self-broadened halfwidth (HWHM) in MHz/Torr at Tref
E (F10.4) = lower state energy in wavenumbers (cm-1)
N (F4.2) = coefficient of temperature dependence of
air-broadened halfwidth
* N_self (F4.2) = coefficient of temperature dependence of
self-broadened halfwidth
* Tref (F7.2) = reference temperature for AGAM and SGAM
* d (F9.7) = shift of transition due to pressure (MHz/Torr)
V1 (I3) = upper state global quanta index
V2 (I3) = lower state global quanta index
Q1 (A9) = upper state local quanta
Q2 (A9) = lower state local quanta
IERS (I1) = accuracy index for S
IERH (I1) = accuracy index for AGAM
* IERN (I1) = accuracy index for N
The asterisks mark entries that are different from HITRAN.
Note that AGAM and SGAM are for the temperature Tref, while S is
still for 296 K!
The molecule numbers are encoded as shown in the table below:
0= Null 1= H2O 2= CO2 3= O3 4= N2O 5= CO
6= CH4 7= O2 8= NO 9= SO2 10= NO2 11= NH3
12= HNO3 13= OH 14= HF 15= HCl 16= HBr 17= HI
18= ClO 19= OCS 20= H2CO 21= HOCl 22= N2 23= HCN
24=CH3Cl 25= H2O2 26= C2H2 27= C2H6 28= PH3 29= COF2
30= SF6 31= H2S 32=HCOOH 33= HO2 34= O 35= CLONO2
36= NO+ 37= Null 38= Null 39= Null 40=H2O_L 41= Null
42= Null 43= OCLO 44= Null 45= Null 46=BRO 47= Null
48= H2SO4 49=CL2O2
All molecule numbers are from HITRAN, except for species with id's
greater or equals 40, which are not included in HITRAN.
(E.g.: For BrO, iso=1 is Br-79-O,iso=2 is Br-81-O.)
The function attempts to read a line of data from the catalogue. It returns false if it succeeds. Otherwise, if eof is reached, it returns true. If an error occurs, a runtime_error is thrown. When the function looks for a data line, comment lines are automatically skipped.
- Parameters
-
is Stream from which to read
- Exceptions
-
runtime_error Some error occured during the read
- Returns
- false=ok (data returned), true=eof (no data returned)
- Date
- 31.10.00
Definition at line 1464 of file linerecord.cc.
References CREATE_OUT0, CREATE_OUT3, extract(), i, iso(), SpeciesRecord::Isotopologue(), QuantumIdentifier::Isotopologue(), max, mf, mqid, my_basic_string< charT >::nelem(), Array< base >::nelem(), Absorption::nelem(), QuantumIdentifier::Species(), global_data::species_data, SPEED_OF_LIGHT, and _CS_basic_sstream_base< _CS_cT, _CS_Tr, _CS_Al >::str().
◆ reduced_magnetic_quadrapole()
|
inline |
Definition at line 435 of file linerecord.h.
References J, N, sqrt(), and wigner6j().
◆ reduced_rovibrational_dipole()
Definition at line 422 of file linerecord.h.
◆ SetAirPressureBroadening()
|
inline |
Definition at line 618 of file linerecord.h.
References LineShape::D0, and LineShape::G0.
◆ SetCutOff()
|
inline |
Definition at line 777 of file linerecord.h.
◆ SetElow()
|
inline |
Definition at line 379 of file linerecord.h.
◆ SetEvlow()
|
inline |
Definition at line 383 of file linerecord.h.
Referenced by nlteSetByQuantumIdentifiers().
◆ SetEvupp()
|
inline |
Definition at line 391 of file linerecord.h.
Referenced by nlteSetByQuantumIdentifiers().
◆ setF()
|
inline |
Set the line center frequency in Hz.
Definition at line 352 of file linerecord.h.
◆ setI0()
|
inline |
Set Intensity.
Definition at line 372 of file linerecord.h.
◆ SetLineMixing2AER()
|
inline |
Definition at line 568 of file linerecord.h.
References i, and ConstVectorView::nelem().
◆ SetLineMixing2SecondOrderData()
|
inline |
Definition at line 561 of file linerecord.h.
References LineShape::LegacyLineMixingData::LM_2NDORDER, and LineShape::LegacyLineMixingData::vector2modellm().
◆ SetLineMixingLimit()
|
inline |
Definition at line 781 of file linerecord.h.
◆ SetLineMixingParameters()
|
inline |
Definition at line 504 of file linerecord.h.
◆ SetLineNormalizationType()
|
inline |
Definition at line 792 of file linerecord.h.
◆ SetLinePopulationType()
|
inline |
Definition at line 805 of file linerecord.h.
Referenced by nlteSetByQuantumIdentifiers().
◆ SetLineShapeModel()
|
inline |
Definition at line 576 of file linerecord.h.
◆ SetLineShapeModelParameter()
|
inline |
Definition at line 597 of file linerecord.h.
References LineShape::SingleModelParameter().
◆ SetLineShapeModelParameters()
|
inline |
Definition at line 591 of file linerecord.h.
References LineShape::string2variable().
◆ SetMirroringType()
|
inline |
Definition at line 785 of file linerecord.h.
◆ SetNLTELowerIndex()
|
inline |
Definition at line 385 of file linerecord.h.
Referenced by nlteSetByQuantumIdentifiers().
◆ SetNLTEUpperIndex()
|
inline |
Definition at line 393 of file linerecord.h.
Referenced by nlteSetByQuantumIdentifiers().
◆ SetPressureBroadeningParameters()
|
inline |
Definition at line 521 of file linerecord.h.
◆ SetPressureBroadeningParametersTemperatureDerivative()
|
inline |
Definition at line 541 of file linerecord.h.
◆ SetQuantumNumberLower() [1/3]
Quantum numbers.
Definition at line 455 of file linerecord.h.
◆ SetQuantumNumberLower() [2/3]
Definition at line 458 of file linerecord.h.
◆ SetQuantumNumberLower() [3/3]
|
inline |
Definition at line 461 of file linerecord.h.
◆ SetQuantumNumberUpper() [1/3]
|
inline |
Definition at line 464 of file linerecord.h.
◆ SetQuantumNumberUpper() [2/3]
Definition at line 467 of file linerecord.h.
◆ SetQuantumNumberUpper() [3/3]
Definition at line 470 of file linerecord.h.
◆ SetSpecial()
|
inline |
Definition at line 611 of file linerecord.h.
◆ SetStandard()
|
inline |
Definition at line 612 of file linerecord.h.
◆ SetVersion5()
|
inline |
Definition at line 305 of file linerecord.h.
◆ SetVersionToLatest()
|
inline |
Definition at line 308 of file linerecord.h.
◆ Sgam()
|
inline |
Self broadened width in Hz/Pa:
Definition at line 401 of file linerecord.h.
◆ Species()
|
inline |
The index of the molecular species that this line belongs to.
The species data can be accessed by species_data[Species()].
Definition at line 312 of file linerecord.h.
◆ SpeciesData()
| const SpeciesRecord & LineRecord::SpeciesData | ( | ) | const |
The matching SpeciesRecord from species_data.
To get at the species data of a LineRecord lr, you can use:
- species_data[lr.Species()]
- lr.SpeciesData()
The only advantages of the latter are that the notation is slightly nicer and that you don't have to declare the external variable species_data.
Definition at line 49 of file linerecord.cc.
References mqid, QuantumIdentifier::Species(), and global_data::species_data.
◆ Ti0()
|
inline |
Reference temperature for I0 in K:
Definition at line 375 of file linerecord.h.
Referenced by operator<<().
◆ UpperQuantumNumber()
|
inline |
Definition at line 485 of file linerecord.h.
◆ UpperQuantumNumbers()
|
inline |
Definition at line 479 of file linerecord.h.
Referenced by operator<<().
◆ UpperStateInQuantumID()
|
inline |
Definition at line 489 of file linerecord.h.
◆ Version()
|
inline |
◆ VersionString()
| String LineRecord::VersionString | ( | ) | const |
Return the version String.
Definition at line 36 of file linerecord.cc.
References mversion, and _CS_basic_sstream_base< _CS_cT, _CS_Tr, _CS_Al >::str().
◆ ZeemanModel() [1/2]
|
inline |
Definition at line 772 of file linerecord.h.
Referenced by operator<<().
◆ ZeemanModel() [2/2]
|
inline |
Definition at line 773 of file linerecord.h.
◆ ZeemanModelInit()
|
inline |
Zeeman Effect Data.
Definition at line 753 of file linerecord.h.
◆ ZeemanModelInitZero()
|
inline |
Definition at line 754 of file linerecord.h.
◆ ZeemanModelLineCount()
|
inline |
Definition at line 767 of file linerecord.h.
References J, and Zeeman::nelem().
◆ ZeemanModelSplitting()
|
inline |
Definition at line 761 of file linerecord.h.
References J.
◆ ZeemanModelStrength()
|
inline |
Definition at line 755 of file linerecord.h.
References J.
Member Data Documentation
◆ ma
|
private |
Definition at line 1175 of file linerecord.h.
◆ mcutoff
|
private |
Cutoff frequency.
Definition at line 1196 of file linerecord.h.
◆ melow
|
private |
Definition at line 1160 of file linerecord.h.
◆ mevlow
|
private |
Definition at line 1163 of file linerecord.h.
◆ mevupp
|
private |
Definition at line 1169 of file linerecord.h.
◆ mf
|
private |
Definition at line 1151 of file linerecord.h.
Referenced by ReadFromHitran2001Stream(), ReadFromHitran2004Stream(), ReadFromLBLRTMStream(), and ReadFromMytran2Stream().
◆ mglower
|
private |
Definition at line 1181 of file linerecord.h.
◆ mgupper
|
private |
Definition at line 1178 of file linerecord.h.
◆ mi0
|
private |
Definition at line 1154 of file linerecord.h.
◆ mlinemixing_limit
|
private |
Line mixing pressure limit.
Definition at line 1199 of file linerecord.h.
◆ mlinenorm
|
private |
Line shape normalization type.
Definition at line 1205 of file linerecord.h.
Referenced by GetLineNormalizationTypeString(), and LineNormalizationTypeFromString().
◆ mlineshapemodel
|
private |
Line function data (pressure broadening and line mixing)
FIXME Richard, please explain.
Definition at line 1187 of file linerecord.h.
◆ mmirroring
|
private |
Line shape mirroring effect type.
Definition at line 1202 of file linerecord.h.
Referenced by GetMirroringTypeString(), and MirroringTypeFromString().
◆ mnlte_lower_index
|
private |
Definition at line 1166 of file linerecord.h.
◆ mnlte_upper_index
|
private |
Definition at line 1172 of file linerecord.h.
◆ mpopulation
|
private |
Line LTE/NLTE type.
Definition at line 1208 of file linerecord.h.
Referenced by GetLinePopulationTypeString(), and LinePopulationTypeFromString().
◆ mqid
|
private |
Definition at line 1148 of file linerecord.h.
Referenced by IsNotSameSpecIso(), IsotopologueData(), Name(), ReadFromHitran2001Stream(), ReadFromHitran2004Stream(), ReadFromLBLRTMStream(), ReadFromMytran2Stream(), and SpeciesData().
◆ mstandard
|
private |
FIXME Richard, please explain.
Definition at line 1190 of file linerecord.h.
◆ mti0
|
private |
Definition at line 1157 of file linerecord.h.
◆ mversion
|
private |
Definition at line 1145 of file linerecord.h.
Referenced by ReadFromArtscat3Stream(), ReadFromArtscat4Stream(), ReadFromArtscat5Stream(), and VersionString().
◆ mzeemanmodel
|
private |
Zeeman effect model class.
Definition at line 1193 of file linerecord.h.
The documentation for this class was generated from the following files:
